IMPRes Usage Tutorial
We prepared two sample datasets for download. You can use them to try our web server or follow the format to create your own submission. One sample dataset is the yeast cell wall damage stress dataset used in our paper (case control). The other one is the yeast high osmolality stress dataset used in our paper (time-series).
How to submit a job
To submit a job, first thing is to specify the organism you are working on.

Then you should upload the seed genes file. The seed genes are the root nodes where the pathways detection process begins. The seed genes file format should be like below....

It contains the ID from KEGG database and String PPI database. If you do not integrate PPI into reference network, the String ID is not required. You can use our gene list conventor tool to convent your gene symbol list to our acceptable format.
Next, you need to provide some information about the target genes. They are the genes that become active during the process and show changes in their expression patterns. You can either upload your own target gene list as the format as seed genes, or you can let us determine target genes automatically based on your target number.
Then you need to upload your expression data. You have to specify the data type of your expression data. If it is a case control type, the format should be a tab seperated .txt file like below...

Be sure to specify how many samples Control and Case have respectively. When it is a time-series data type, the format should be like below...
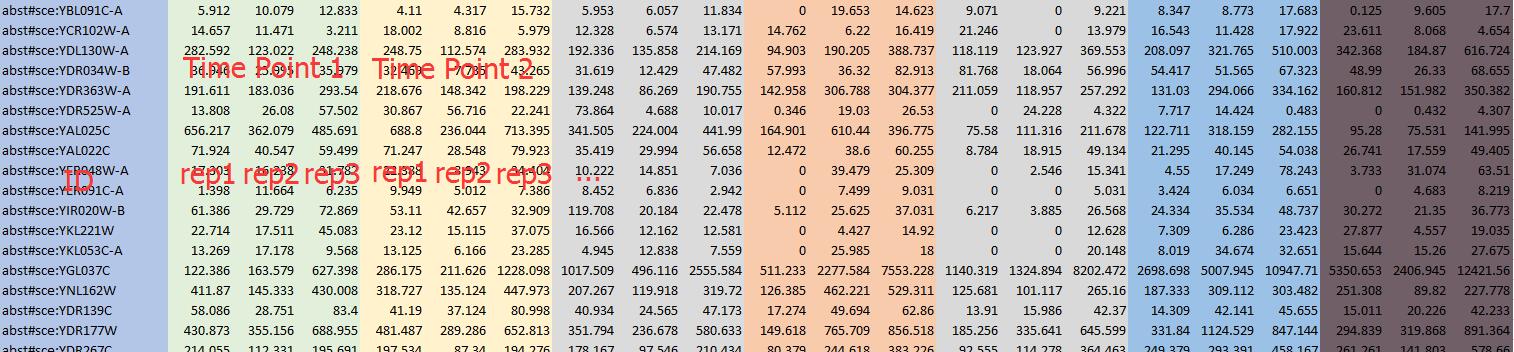
Be sure to specify how many time points your data has and how many replicates each time point has. We have provide the Gene expression conventor to help you convert you expression data to our acceptable format.
Finally, you can decide if integrating PPI data into reference network. Note: integrating PPI data would make the calculating time increase dramaticaly, since the interactions in String database is much more than the interactions in KEGG databse.
